Research Programme: Function of the p53 family member p73
Principal Investigator: Prof. Gerry Melino

Understanding the molecular mechanisms of Cancer is a major challenge for therapy.
The scientific interests of Professor Melino are focused on Programmed Cell Death or apoptosis, in neural and epidermal models. Originally, he worked on the molecular mechanisms of cell death in the skin, a process known as cornification or formation of the cornified envelope. The molecular events were investigated in vitro and in animal models as well as in human genetic pathologies. The role of transglutaminases (type 1, 2, 3, and 5) and their substrates (SPRs, loricrin, keratins) were investigated at biochemical and genetic levels. While still keeping an interest on these aspects, his current work is focused on the family members of the tumour suppressor p53, namely p73 and p63. Indeed, the molecular events driven by DNA damage to elicit the function of p63/p73 and their transcriptional regulation, is investigated in vitro, as well as through the development of several transgenic mice and knockout work for p63 or p73. He identified 8 splicing isoforms of p73, its involvement in DNA damage response, transcriptional regulation, degradation pathways, and mechanisms of killing. His group has developed selective knockout mouse models, allowing the understanding of their neuronal phenotype, as well as female infertility.
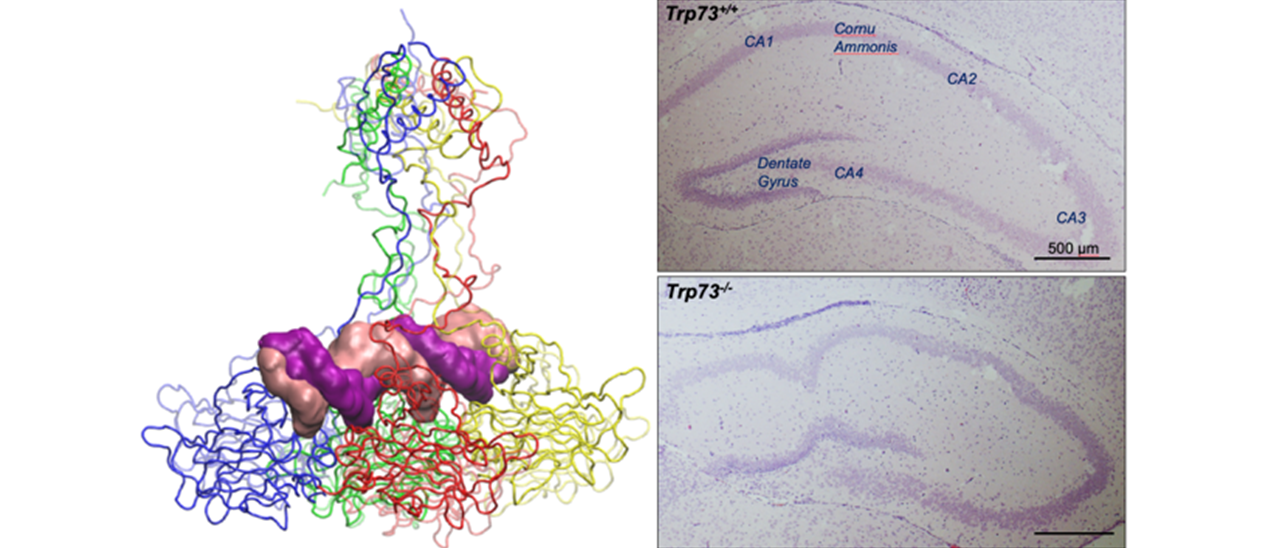
The TAp73 isoforms display tumour suppressive functions by triggering cell cycle arrest and apoptosis in response to genotoxic stress. As a result, TAp73-specific knockout mice have a high occurrence of spontaneous and carcinogen-induced tumours. The C-terminal segment of the longest p73 isoforms (p73α) possesses a putative protein-protein interaction motif, the SAM domain. At present, the FUNCTIONAL RELEVANCE of the SAM domain and the specific functional contribution of the p73α isoforms to neurogenesis and tumorigenesis remain unknown. Whilst several p73 target gene have been identified and characterized so far, our knowledge on the p73 binding proteins is quite limited. We have recently defined a comprehensive p73α INTERACTOME. The identification of proteins that associate with p73α may provide a clue for better understanding the molecular mechanisms regulating the specific functions of this isoform and its involvement in tumorigenesis. In addition, to further explore the functional implications of p73α we have recently generated a selective p73α KNOCKOUT MOUSE MODEL. The long-term goal of the current proposal is to identify and characterize protein-binding partners of p73 and to study the functional outcome of these associations on its tumour suppressive properties. Furthermore, by exploiting a conditional p73α defective mouse, we seek to characterize the in vivo role of the full-length p73 isoform. The ongoing research is accomplished through the following tasks:
TASK 1. Identification of novel binding partners of p73α. We aim to biochemically and functionally validate the association of p73α with several potential protein interactors, that we have identified by using the BioID approach coupled with mass spectrometry.
TASK 2. Role of the carboxy-terminus of p73 in neurodevelopment and tumorigenesis. By exploiting a selective p73α knockout mouse model, we aim to investigate the functional contribution of the p73α isoforms. Particular emphasis will be given to unveil a role for the SAM domain in mediating the tumour suppressive functions of p73.
Due to the high structural complexity exhibited by the p73 proteins, it has been difficult to assign specific functions to each protein variant. Information gained from our studies is expected to be of direct RELEVANCE TO CANCER BIOLOGY, because it will foster our understanding of the role of TAp73α in cancer and in neurobiology. The validation of TAp73 as a tumour suppressor may offer novel therapeutic approaches to enhance the chemosensitivity of cancer cells harbouring mutated or inactivated p53.